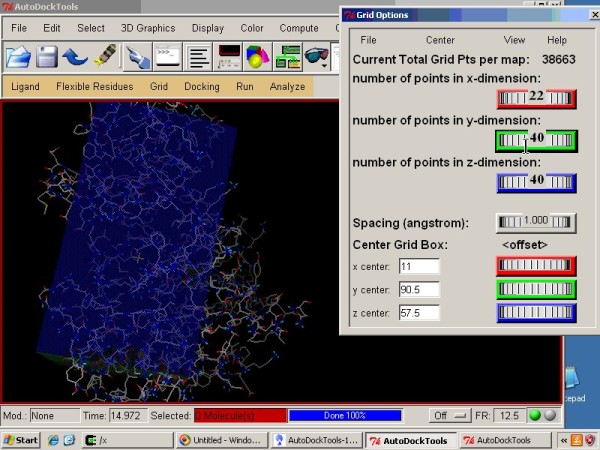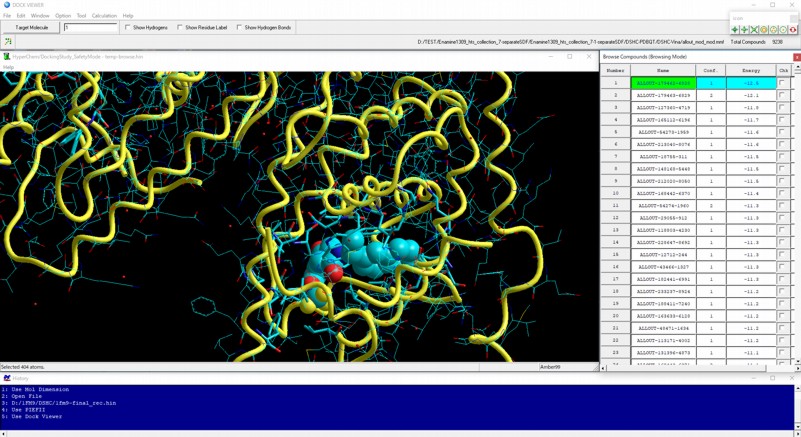
AutoDock Vina In Silico Screenings Interface: Docking Study with HyperChem - Institute of Molecular Function -
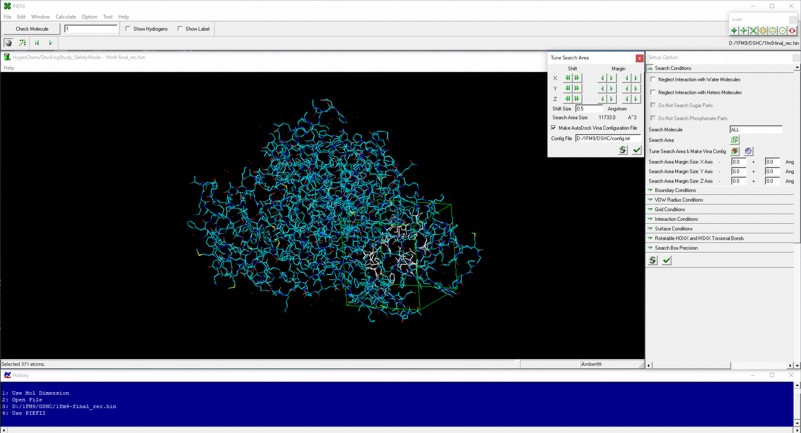
AutoDock Vina In Silico Screenings Interface: Docking Study with HyperChem - Institute of Molecular Function -
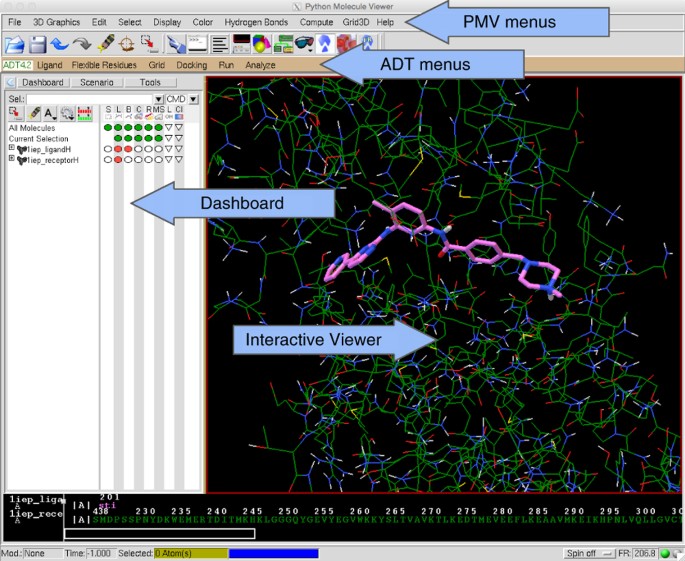
Computational protein–ligand docking and virtual drug screening with the AutoDock suite | Nature Protocols

Optimization of Autodock vina program by docking the co-crystal ligand... | Download Scientific Diagram

Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock - ScienceDirect

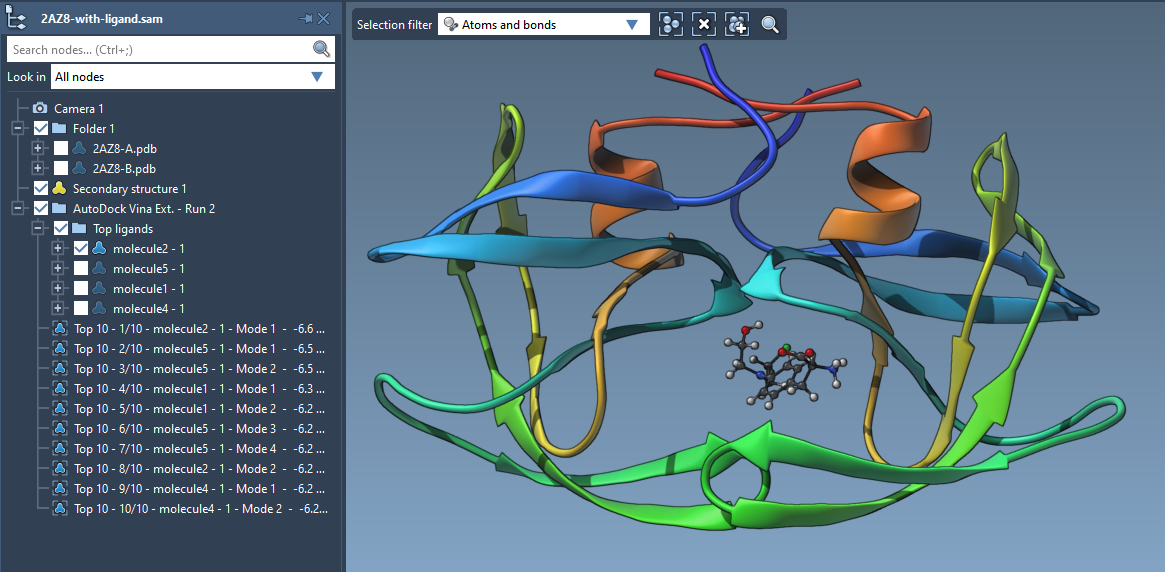
The new version of AutoDock Vina Extended is out for SAMSON 2020 R2 | The new version of AutoDock Vina Extended includes interactive analysis of docking results by linking plots to conformations.
A) The output of AutoDock Vina showing the binding site residues of... | Download Scientific Diagram
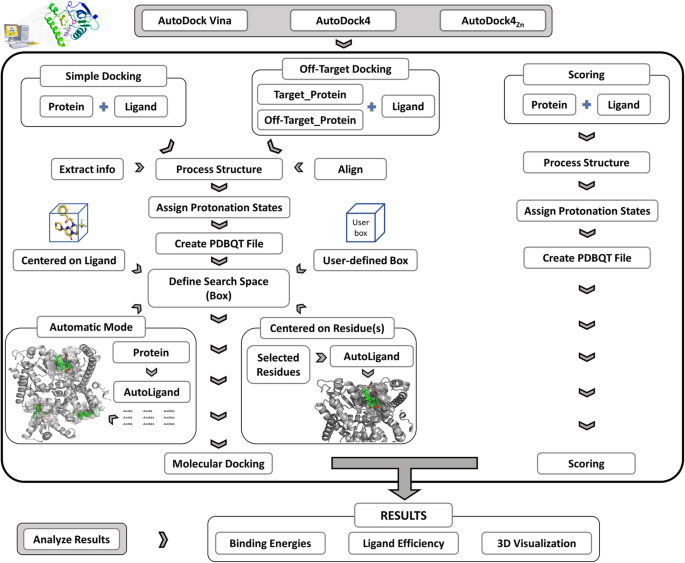
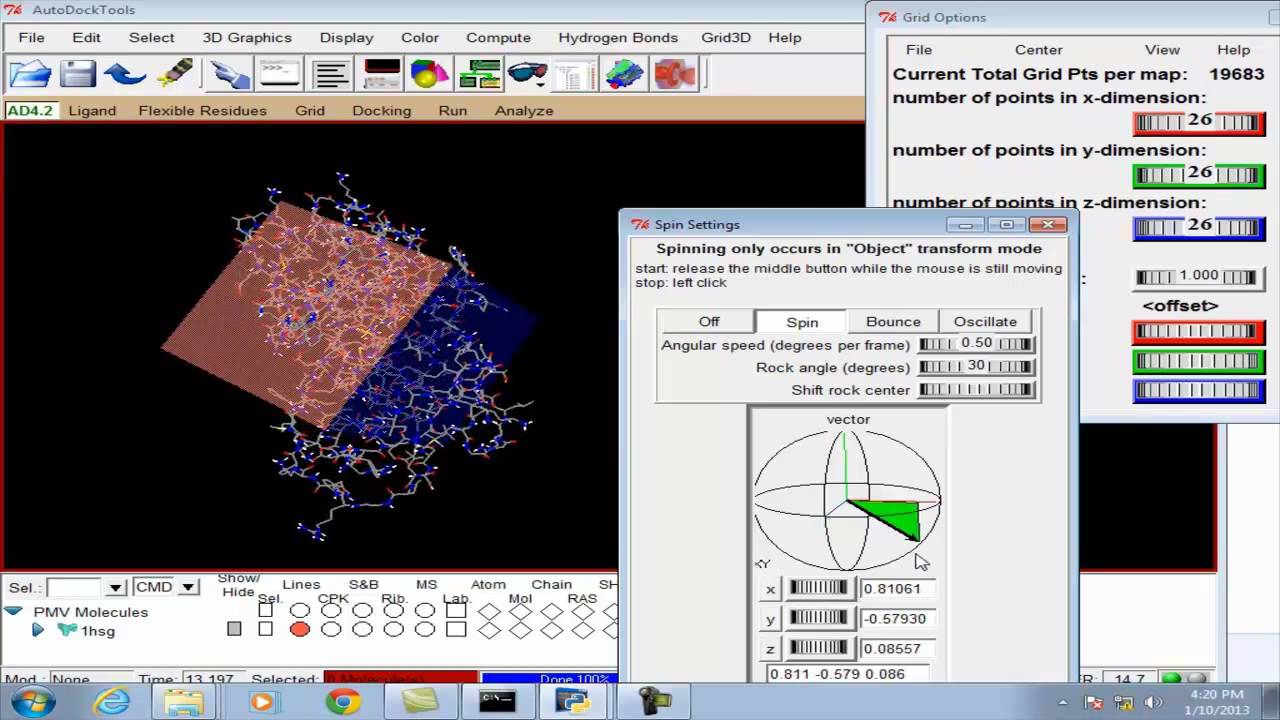
AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4 | Biology Direct | Full Text